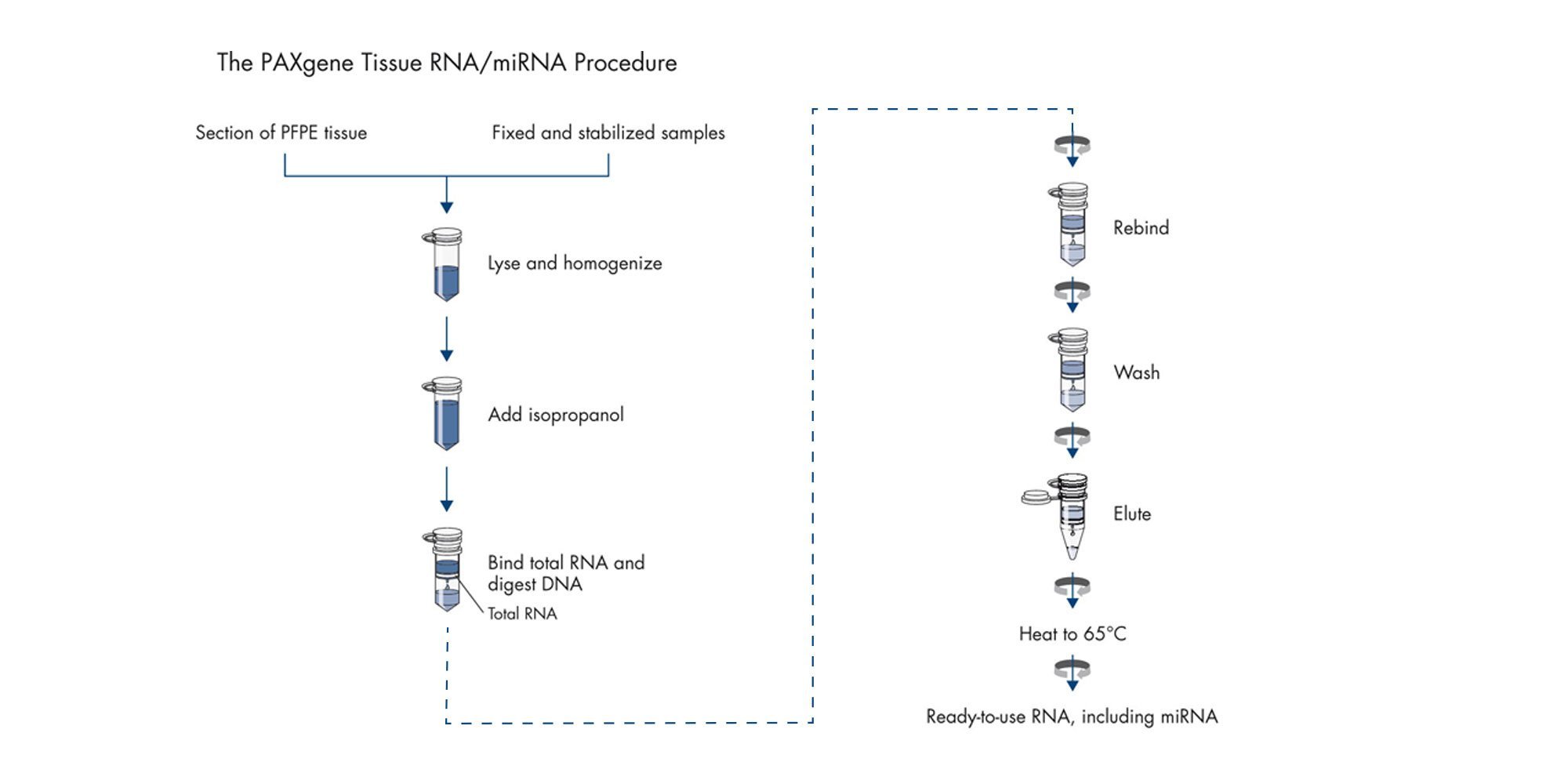
PAXgene Tissue RNA/miRNA Kit
Purification and quality of biomolecules from PAXgene Tissue-treated samples
1. Is it possible to use formalin-fixed, paraffin-embedded (FFPE) kits and protocols to isolate biomolecules from PAXgene Tissue-fixed, paraffin-embedded (PFPE) tissues?
No. Procedures developed for the extraction of biomolecules from FFPE tissues include prolonged proteinase K digestion and heating steps to remove chemical modifications introduced by formalin. Since the PAXgene Tissue System does not chemically modify biomolecules, these steps are not necessary and, in fact, lead to degradation of biomolecules. Instead, use dedicated PAXgene Tissue Kits and supplementary protocols for extraction of biomolecules from PAXgene Tissue-treated samples.
2. What is the purity of nucleic acids extracted with the PAXgene Tissue Kits?
The PAXgene Tissue DNA and RNA/miRNA Kits are based on proven QIAGEN technologies. Nucleic acids isolated with these kits are generally of high purity.
On average, measurements of the A260/A280 ratio for DNA purified with the PAXgene Tissue DNA Kit are >1.7, and ratios for RNA, including miRNA, purified with the PAXgene Tissue RNA/miRNA Kit are >1.8.
3. What is the average RNA yield, including miRNA, from PAXgene Tissue-fixed, paraffin-embedded (PFPE) tissues?
RNA yield, including miRNA, depends on several parameters, such as tissue type, time from resection until fixation, fixation time, processing protocol used and age and storage conditions of the PFPE block.
In a study with PFPE tissue sections (area: 100 mm²; thickness: 10 µm) median RNA yield from rat liver was 4.2 µg (n=58), from kidney 2.2 µg (n=58), from spleen 4.7 µg (n=58), from intestine 4.7 µg (n=58) and from lung 0.9 µg (n=58). See the Technical Note "Yield, purity, and integrity of RNA purified from PAXgene Tissue fixed, paraffin-embedded (PFPE) rat tissue" under Resources.
4. How well is RNA integrity preserved in PAXgene Tissue-fixed, paraffin-embedded (PFPE) tissues?
Similar to yield, RNA integrity depends on several parameters, such as tissue type, time from resection until fixation, fixation time, processing protocol and age and storage conditions of the PFPE block. For examples of RNA integrity values from rat tissues under ideal workflow conditions, see Groelz et al. 2013under References. For examples of RNA integrity from clinical samples, see Viertler et al. 2012.
5. What are the yield and integrity of nucleic acids isolated from blocks of PAXgene Tissue-fixed, cryo-embedded (PFCE) tissues?
DNA and RNA isolated from PFCE tissue specimens are of high quantity and quality, comparable to PFPE tissue.
6. Are special kits and protocols required for the isolation of biomolecules from PAXgene Tissue fixed, cryo-embedded (PFCE) tissues?
No. Regular PAXgene Tissue Kits can be used for the isolation of RNA, miRNA and DNA from PFCE tissue. Supplementary protocols developed specifically for the extraction of biomolecules from PFCE samples are available under Resources.
7. Which kits and protocols can be used for the isolation of nucleic acids from microdissected PAXgene-Tissue fixed, paraffin-embedded (PFPE) and PAXgene Tissue-fixed, cryo-embedded (PFCE) specimens?
Regular PAXgene Tissue Kits can be used for the isolation of RNA, miRNA and DNA from microdissected PFPE and PFCE tissue. Supplementary protocols developed specifically for the extraction of biomolecules from microdissected PFPE and PFCE samples are available under Resources.
Molecular analysis of biomolecules purified from PAXgene Tissue-treated samples
1. What is the RT-PCR performance of RNA purified from PAXgene Tissue-fixed, paraffin-embedded (PFPE) and PAXgene Tissue-fixed, cryo-embedded (PFCE) tissues compared to RNA from snap-frozen or formalin-fixed, paraffin-embedded (FFPE) tissues?
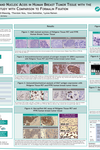
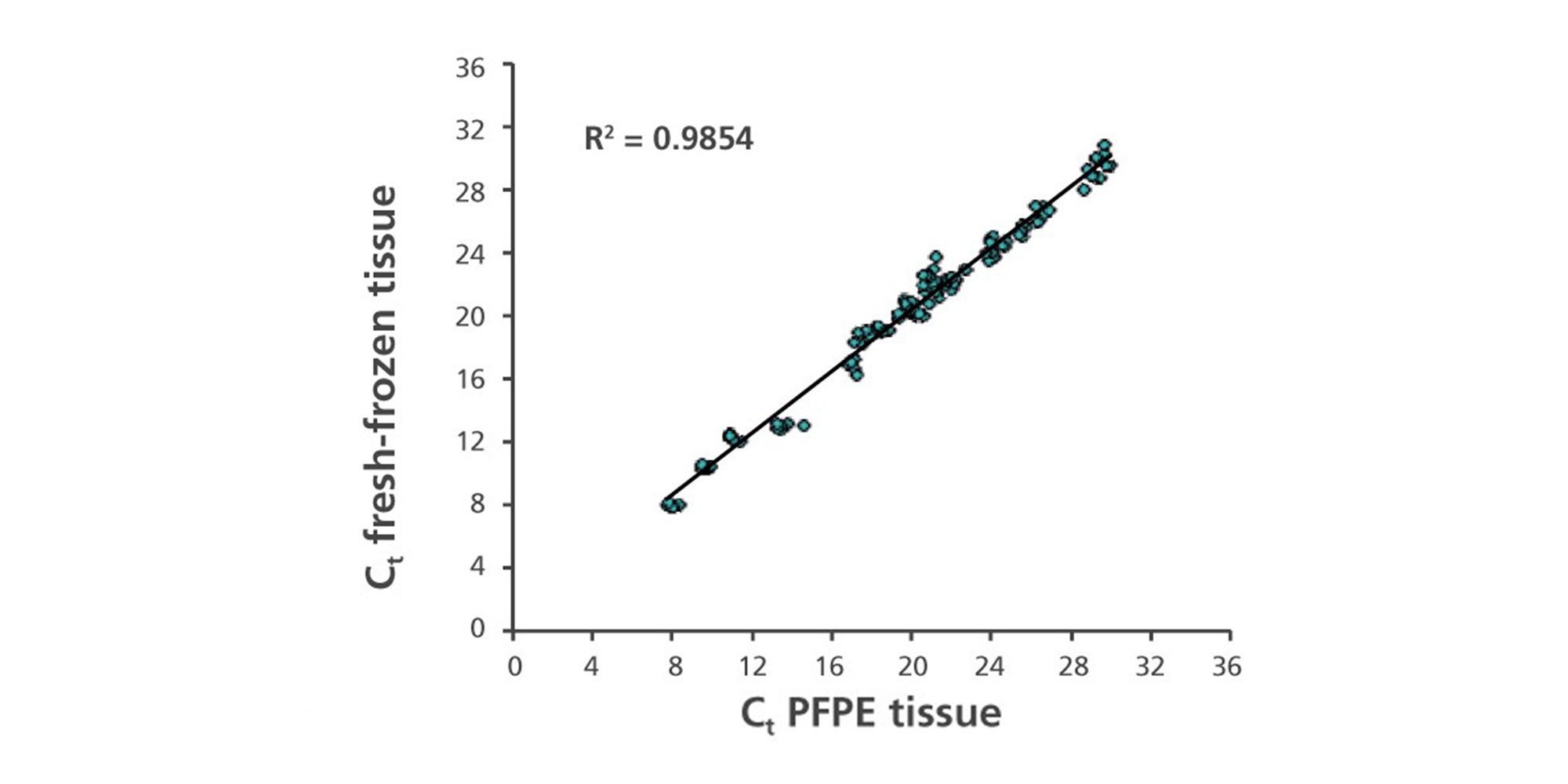
RNA, including miRNA, purified from PFPE and PFCE is free of chemical modifications and performs similarly or identically to RNA isolated from frozen tissue. For examples of the correlation of gene expression levels in snap-frozen tissue, FFPE and PFPE, see Figure 4 in Groelz et al. 2013and Figure 3 in Viertler et al. 2012 under Resources.
![Figure 2. RNA purified without chemical modification from PFPE tissue using the PAXgene Tissue RNA/miRNA Kit SYBR Green real-time RT-qPCR was performed with 10 ng RNA from cryopreserved, formalin-fixed, paraffin-embedded (FFPE) or PAXgene Tissue-fixed, paraffin-embedded (PFPE) rat tissue (modified according to Groelz et al. (2013) Non-formalin fixative versus formalin-fixed tissue: a comparison of histology and RNA quality. Exp. Mol. Path. 94, 188). Depicted are the average delta-Ct values (delta-Ct = Ct[FFPE] – Ct[cryo] or delta-Ct = Ct[PFPE] – Ct[cryo]) from 6 different assays with amplicons ranging from 109 to 465 bp.](/storage/images/Content/Product_pages/Tissue/RNA_miRNA/SYBR_Green_real-tome_RT-qPCR_FFPE_PFPE_2.jpg)